Workflow and applications
The unique industry leading 1-stage workflow was co-developed with industry leaders in agricultural biotechnology to ensure it is optimised for industrial throughput and simplified to enable lab efficiency.
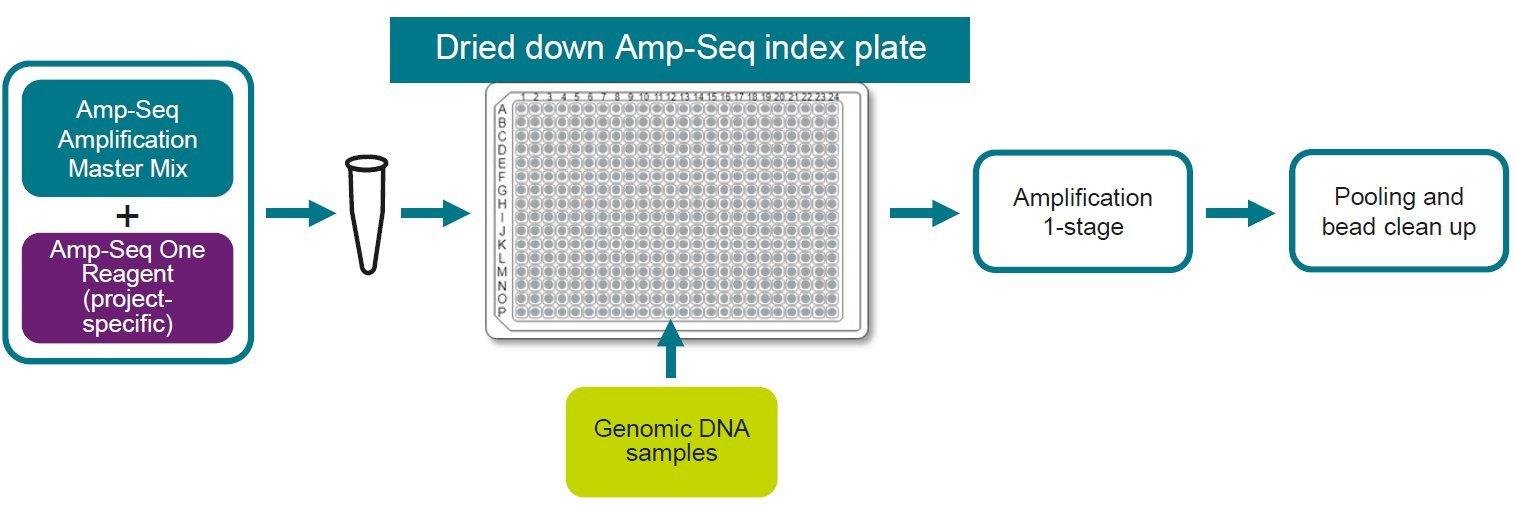
Amp-Seq One has been developed for high-throughput genotyping applications including:
- Seed QC
- Genomic selection
- Parentage testing
- Marker assisted selection (MAS) and back-crossing (MABC)
- Other high-throughput genotyping applications
Custom targeting oligo libraries
We work with you to design a custom panel that meets your genotyping goals. With our proprietary design software, we design high-quality panels that result in high sequencing efficiency and typically have very high SNP call rates and evenness of coverage. These panels can be further refined and optimised for high priority markers.
A design service is available for custom targeting oligo panels. To initiate a custom panel design project, please contact your LGC Biosearch Technologies representative.
The design process includes custom panel design, panel production (per marker), panel validation and optimization (optional, recommended) and a test kit for in-house testing.
| Description |
Part code |
Explanation |
| Custom panel design (in-silico) |
GEN-9910-001 |
In-silico design and optimization of a new Amp-Seq One panel. Does not require commitment for panel production. |
| Panel production (per marker) |
GEN-9910-002 |
Only charged for markers that pass approval during the design phase. Can be ordered separately from design. |
| Panel validation & optimization |
GEN-9910-003 |
Recommended for all new designs. Includes wet-lab validation and optimization on 384 samples (extraction, library preparation, sequencing, analysis) |
| Custom design test kit |
GEN-9910-004 |
Recommended for new Amp-Seq One users. Trial kit for in-house testing, including all required reagents (excl. template DNA) |
Following the collaborative design process, your project manager will provide you with a design specific ‘Design ID’ number. Upon ordering your kits, the project specific targeting oligo library is added to your kit by your Biosearch Technologies representative. You will be contacted to confirm the ‘Design ID’ number for the desired targeting oligo library for the kit you have ordered
Online ordering: When ordering a kit with a custom panel online, please ensure to leave your Amp-Seq One targeting oligo design ID number (provided by your project manager) in the notes section upon checkout to ensure we add the correct panel to your kit.
Industry standard pre-designed panels
Amp-Seq One kits with pre-designed panels are available for fast, easy and cost-efficient validation of the technology and for production scale screening applications. These pre-designed panel kits do not require design, production, validation and optimization.
| Description |
Part code |
Explanation |
| Pre-designed panel kit |
GEN-9910-005 |
Alternative to kits with custom designs. Pre-designed panel kits do not require design, production or validation. |
Industry standard pre-designed Amp-Seq panels are currently available for bovine and maize, additional panels are in development. For more information about pre-designed panels or to develop your own custom panel, please contact your LGC representative.
Core reagent kits
Generic part codes for Amp-Seq One core reagent kits and index plates are in the table below.
| Description |
Part code |
| Amp-Seq One kit incl. targeting oligo library (384 rxn) |
GEN-9910-384 |
| Amp-Seq One kit incl. targeting oligo library (3840 rxn) |
GEN-9910-3840 |
| LGC Nexar - Amp-Seq One kit incl. targeting oligo library (384 rxn) |
GEN-9910-384 |
| LGC Nexar - Amp-Seq One kit incl. targeting oligo library (3840 rxn) |
GEN-9910-3840 |
| Amp-Seq One dried down RT storage CDI (indexing) plate 1-96 |
GEN-9902-001 to -096 |
The kits are available in 384 and 3,840 reaction sizes that include sufficient coverage to accommodate the use of liquid handlers.
To order reagents for more than 3,840 reactions please order multiples of the relevant kits, for example: for processing 8,000 samples, please order 2 x 3,840 rxn kits and 1 x 384 kit. To determine the number of samples you can multiplex depending on your sequencing instrument and flow-cell type, please refer to the Amp-Seq One manual for an overview of multiplexing options on various sequencing instruments.
Online ordering: After selecting the desired kit format and number, please select the desired index plates from the “kit items” section.
Automation and implementation support
Amp-Seq One is compatible with most liquid handling systems, see technical specifications for details. Additionally, LGC can support implementation on your workflow.
Additional services include:
- Optimisation for specific input material (species and sample types)
- Wet-lab implementation support
- Automation compatibility and scale up support
- BiosearchCaller data analysis pipeline installation and support
- Customisation of wet-lab chemistry, automation and analysis pipeline
Data analysis
The BiosearchCaller analysis pipeline is specifically optimized for genotype calling in Amp-Seq and Amp-Seq One, and is available to Amp-Seq customers without a license. The pipeline is flexible and can be configured to suit your requirements, handling multiple species and panels. The BiosearchCaller calls genotypes by considering read alignments after quality filtering, allele balance, and local haplotype structure, utilizing Bayesian models to detect polymorphisms from Amp-Seq sequencing data, thereby enabling the accurate detection of SNPs and indels. The BiosearchCaller genotypes are reported in multiple formats to facilitate downstream analysis, along with additional metrics that support troubleshooting.