Competent cells for transformation
- Home
- Competent cells for transformation
- Protein Expression
- OverExpress Electrocompetent Cells
OverExpress Electrocompetent Cells
Product Details
Show HideEach OverExpress kit contains Electrocompetent Cells in SOLO packaging (1 transformation per tube), Expression Recovery Medium (lactose minus), pUC19 Positive Control Plasmid, pAVD10 Verification Plasmid, and complete protocols.
OverExpress Electrocompetent Cells
Key features
Show Hide- Express genes cloned into any T7 vector with these BL21(DE3) derivatives
- Effective in expressing toxic membrane proteins
- Cited in over 350 research articles
- Recommended for membrane proteins with T7 vector
Cannot add to favourites
An option must be selected in order to add to favourites
Similar Products
Phage Display Library Construction
ER2738 Electrocompetent Cells
Low Endoxin Cells
ClearColi BL21(DE3) Electrocompetent Cells
Large or Difficult Cloning
TransforMax EPI300-T1R Electrocompetent Cells
Phage Display Library Construction
TG1 Electrocompetent Cells
Phage Display Library Construction
MC1061 F-Electrocompetent Cells
Phage Display Library Construction
SS320 (MC1061 F') Electrocompetent Cells
Competent cells for transformation
TransforMax EC100D Electrocompetent E. coli
Phage Display Library Construction
ER2738 Electrocompetent Cells
Low Endoxin Cells
ClearColi BL21(DE3) Electrocompetent Cells
Large or Difficult Cloning
TransforMax EPI300-T1R Electrocompetent Cells
Phage Display Library Construction
TG1 Electrocompetent Cells
Phage Display Library Construction
MC1061 F-Electrocompetent Cells
Phage Display Library Construction
SS320 (MC1061 F') Electrocompetent Cells
Competent cells for transformation
TransforMax EC100D Electrocompetent E. coli
Phage Display Library Construction
ER2738 Electrocompetent Cells
Product information
E. coli BL21(DE3) strains, like E. cloni™ EXPRESS Competent Cells provide reliable expression of many genes cloned into T7 expression vectors (e.g.., pET or pSMART™-cDNA vectors). However, in some cases expression is minimal or not detectable because the recombinant protein, when expressed, is deleterious or lethal to these standard BL21 strains. Examples of such toxic proteins include many membrane proteins, some cytoplasmic proteins, and nucleases. Unfortunately, successful expression of one or more toxic proteins is often important to the experimental goal.
OverExpress Electrocompetent and Chemically Competent Cells are E. coli strains that are effective in expressing toxic proteins from all classes of organisms, including eubacteria, yeasts, plants, viruses, and mammals. The effectiveness of these new strains in expressing toxic proteins has been validated in more than 350 publications.
The OverExpress strains contain genetic mutations phenotypically selected for conferring tolerance to toxic proteins. The strain C41(DE3) was derived from BL21(DE3). This strain has at least one mutation, which prevents cell death associated with expression of many recombinant toxic proteins. The strain C43(DE3) was derived from C41(DE3) by selecting for resistance to a different toxic protein and can express a different set of toxic proteins to C41(DE3). Figure. 1 graphically illustrates the advantages of the OverExpress Competent Cells, compared to standard BL21(DE3) cells, in expressing toxic proteins.
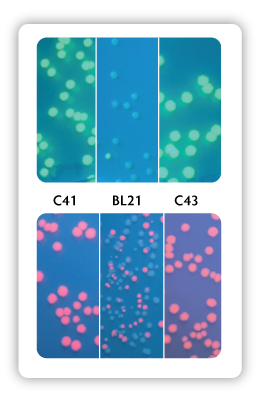
Figure 1. Green Fluorescent Protein (top) or Red Fluorescent Protein (bottom) expressed from a T7 promoter construct that was transformed into C41, BL21, or C43 competent cells spread on IPTG plates to induce protein expression.
Table. 1 and Figure. 2 summarise transformation effectiveness, tolerance of expression-induced toxicity, and protein expression for T7 expression plasmids coding for a variety of recombinant proteins. These results demonstrate that the OverExpress C41(DE3) and C43(DE3) strains are clearly superior to the parental BL21(DE3) in transformation and expression of toxic proteins.
Table 1. Comparison of OverExpress C41(DE3) and C43(DE3) cells with the parental strain BL21(DE3) in transformation and expression of heterologous proteins.**
| Strain |
Transformation
Success Ratea |
Expression-induced Toxicityb
|
Expressing Plasmidsc
|
|
BL21(DE3)
|
16/26 (62%)
|
25/26 (96%)
|
14/26 (54%)
|
|
C41(DE3)
|
28/28 (100%)
|
14/28 (50%)
|
24/28 (86%)
|
|
C43(DE3)
|
28/28 (100%)
|
1/28 (4%)
|
23/28 (81%)
|
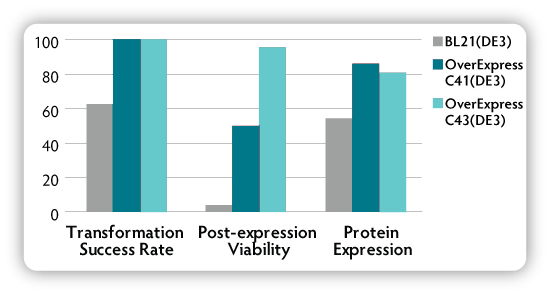
Figure 2. Comparison of OverExpress C41(DE3) and C43(DE3) cells with the parental strain BL21(DE3) in transformation and expression of heterologous proteins.**
a Transformation success corresponds to the presence of colonies on LB+ampicillin agar following transformation with a plasmid.
b Expression toxicity corresponds to the absence of colonies on LB+ampicillin+IPTG agar following transformation with a plasmid.
c Expressing plasmids corresponds to observation of a heterologous protein in the total cell pellet on Coomassie-stained SDS-PAGE following growth of a colony in LB+ampicillin medium and induction with IPTG.
**L. Dumon-Seignovert, G. Cariot, and L. Vuillard (2004). Protein Expression and Purification 37, 203-206. Data used with permission.
As in standard BL21(DE3) strains, OverExpress C41(DE3), C41(DE3)pLysS, C43(DE3), and C43 (DE3)pLysS are lysogens of λDE3. These strains carry a chromosomal copy of the T7 RNA Polymerase gene under the control of the lacUV5 promoter. These strains are suitable for production of protein from target genes cloned into T7-driven expression vectors. OverExpress C41(DE3), C41(DE3) pLysS, C43(DE3), and C43(DE3) pLysS are also deficient in the lon and ompT proteases.
OverExpress C41(DE3)pLysS and C43(DE3)pLysS also carry a chloramphenicol-resistant plasmid that encodes T7 lysozyme, which is a natural inhibitor of T7 RNA polymerase. Cells containing pLysS produce a small amount of T7 lysozyme. These strains are used to suppress basal expression of T7 RNA polymerase prior to induction, thus stabilising recombinants encoding particularly toxic proteins.
Related Products
Size
E. cloni 10G Chemically Competent Cells
Size
E. cloni EXPRESS Competent Cells
General Cloning and Library Construction
HI-Control 10G Chemically Competent Cells
Protein Expression
HI-Control BL21(DE3) Chemically Competent Cells
Protein Expression
OverExpress Chemically Competent Cells
General Cloning and Library Construction
E. cloni 5-alpha Chemically Competent Cells
Large or Difficult Cloning
CopyCutter EPI400 Competent Cells
Format
Endura Competent Cells
Format
TransforMax EC100 Competent Cells
Size
E. cloni 10G Chemically Competent Cells
Size
E. cloni EXPRESS Competent Cells
General Cloning and Library Construction
HI-Control 10G Chemically Competent Cells
Protein Expression
HI-Control BL21(DE3) Chemically Competent Cells
Protein Expression
OverExpress Chemically Competent Cells
General Cloning and Library Construction
E. cloni 5-alpha Chemically Competent Cells
Large or Difficult Cloning
CopyCutter EPI400 Competent Cells
Format
Endura Competent Cells
Format
TransforMax EC100 Competent Cells
Size
E. cloni 10G Chemically Competent Cells
Size
E. cloni EXPRESS Competent Cells
OverExpress Genotypes
OverExpress C41(DE3): F – ompT hsdSB (rB- mB-) gal dcm (DE3)
OverExpress C41(DE3)pLysS: F – ompT hsdSB (rB- mB-) gal dcm (DE3) pLysS (CmR)
OverExpress C43(DE3): F – ompT hsdSB (rB- mB-) gal dcm (DE3)
OverExpress C43(DE3)pLysS: F – ompT hsdSB (rB- mB-) gal dcm (DE3) pLysS (CmR)
Table 1. OverExpress Transformation Efficiencies
| Electrocompetent Cells | cfu/µg DNA |
| C41(DE3) | > 1 × 109 |
| C43(DE3) | > 1 × 109 |
| Chemically Competent Cells | |
| C41(DE3) | > 1 × 106 |
| C41(DE3)pLysS | > 1 × 106 |
| C43(DE3) | > 1 × 106 |
| C43(DE3)pLysS | > 1 × 106 |
SDS
Manuals and user guides
Product information sheets
You may also be interested in...
Large or Difficult Cloning
CopyCutter Induction Solution
Size
TransforMax EPI300
Large or Difficult Cloning
CopyControl Induction Solution
Recovery Media
Expression Recovery Medium
Large or Difficult Cloning
CopyCutter Induction Solution
Size
TransforMax EPI300
Large or Difficult Cloning
CopyControl Induction Solution
Recovery Media
Expression Recovery Medium
Large or Difficult Cloning
CopyCutter Induction Solution
Size
TransforMax EPI300
Access support
Need some support with placing an order, setting up an account, or finding the right protocol?
Contact us