NxSeq™ Single-cell RNA-seq beads contain a covalently attached library of barcoded oligo-dT capture oligonucleotides to enable single-cell mRNA sequencing experiments using microfluidics and well-based instruments, or similar. Every bead-attached oligo contains a 12 base, bead-specific barcode, a 14 base unique molecular identifier (UMI) that is distinct between oligos, and a 3’ 30 base, oligo-dT mRNA capture sequence.
Oligo sequences on each bead:
Bead – Linker – 5’ - TTTTTTTAAGCAGTGGTATCAACGCAGAGTAC JJJJJJJJJJJJ NNNNNNNNNNNNNV T 30 - 3’
J : 12 base bead barcode – same on every oligo on a bead, distinct between beads
N : first 13 bases of the UMI (dA, dC, dG, or dT) - varies across oligos on each bead
V : last non-dT base of the 14 base UMI – varies across oligos on each bead
T 30 : 30 base oligo-dT capture sequence
The NxSeq Single-cell RNA-seq Beads are built using a hard, size selected hydroxylated methacrylic polymer bead and undergo extensive quality control testing to ensure consistent RNA-sequencing results day-to-day and lot-to-lot. These beads are carefully size selected to produce beads with a mean size of 34 µm with less than a 5% fragmentation rate. Post-production, the beads are tested by next gen sequencing to verify minimal errors in oligo synthesis, well-balanced UMIs, and correct oligo length on the beads. These beads are also functionally tested in a qPCR-based, “mRNA” capture assay using synthetic polyA RNA-DNA hybrid oligo (Figure. 4) to carefully monitor their performance characteristics.
Taken together, you can rest easy knowing that these beads will perform consistently in your demanding single-cell RNA sequencing experiments.
If you’re interested in bulk quantities of these or similar custom beads, please contact us.
Classic, single-cell RNA sequencing bead design of the NxSeq Beads
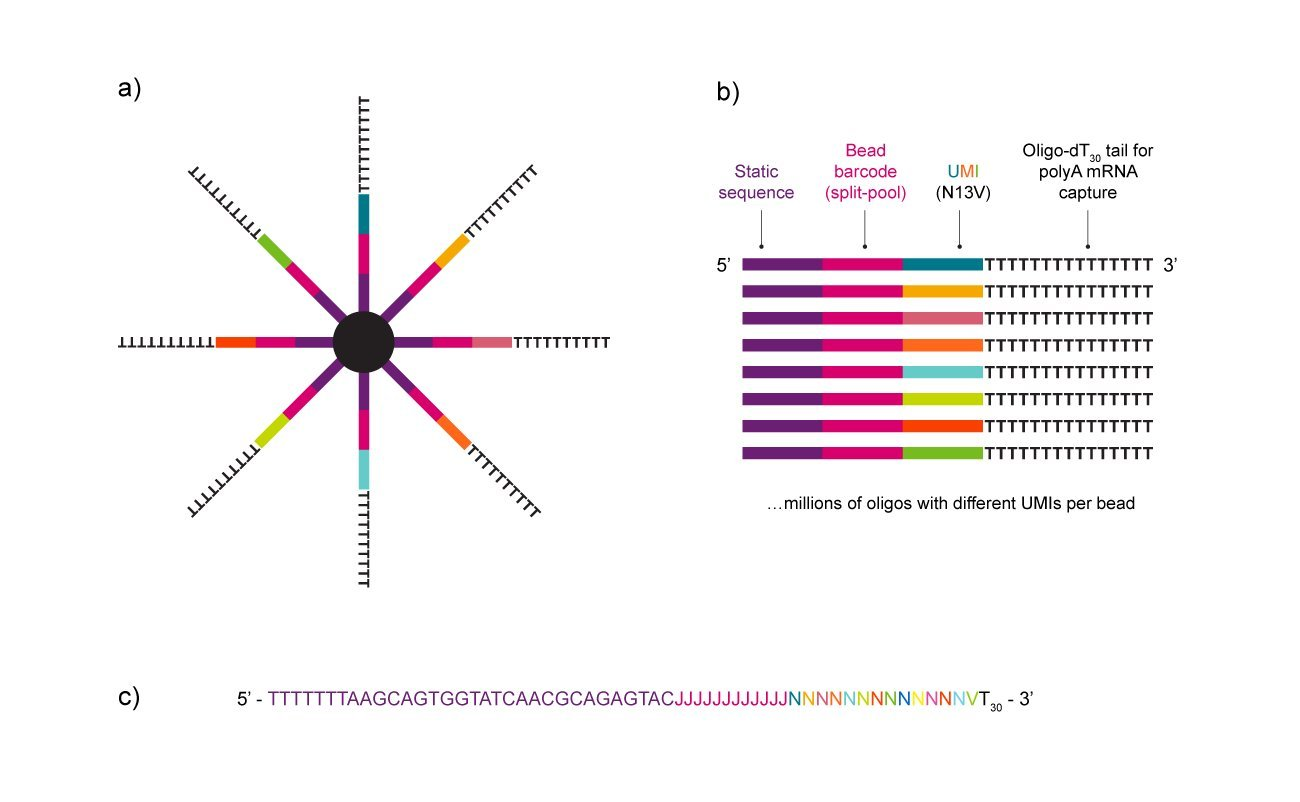
Figure 1. Design and key components of the NxSeq Single-cell RNA-seq Beads. (a) Graphical illustration showing oligos attached to each bead. (b) Detailed look at the parts of each oligo attached to each bead. Note that per bead, only the UMI sequences vary. (c) Actual sequence of oligos attached to each bead. Note that the J bases correspond to the bead barcode and are constant per bead but different from bead to bead. The N13V bases correspond to the UMIs which are different across all oligos per bead.
High quality, size selected beads with minimal fragmentation
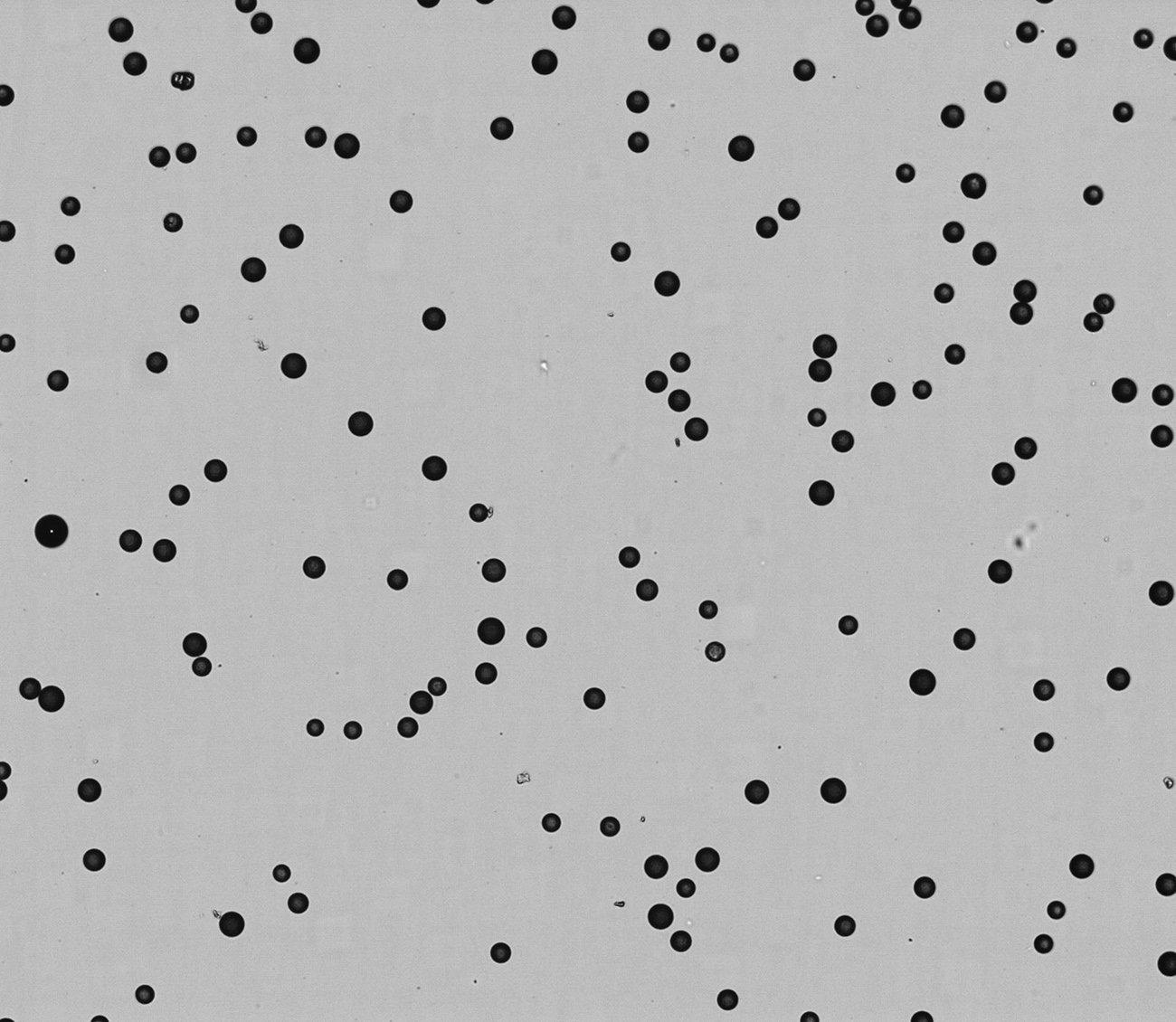
Figure 2. Image of NxSeq Single-cell RNA-seq Beads showing bead size and lack of fragments. Mean size of the NxSeq Beads is 34 µm +/- 2 µm with less than 5% fragmentation rate.
Excellent on-bead oligo sequence characteristics
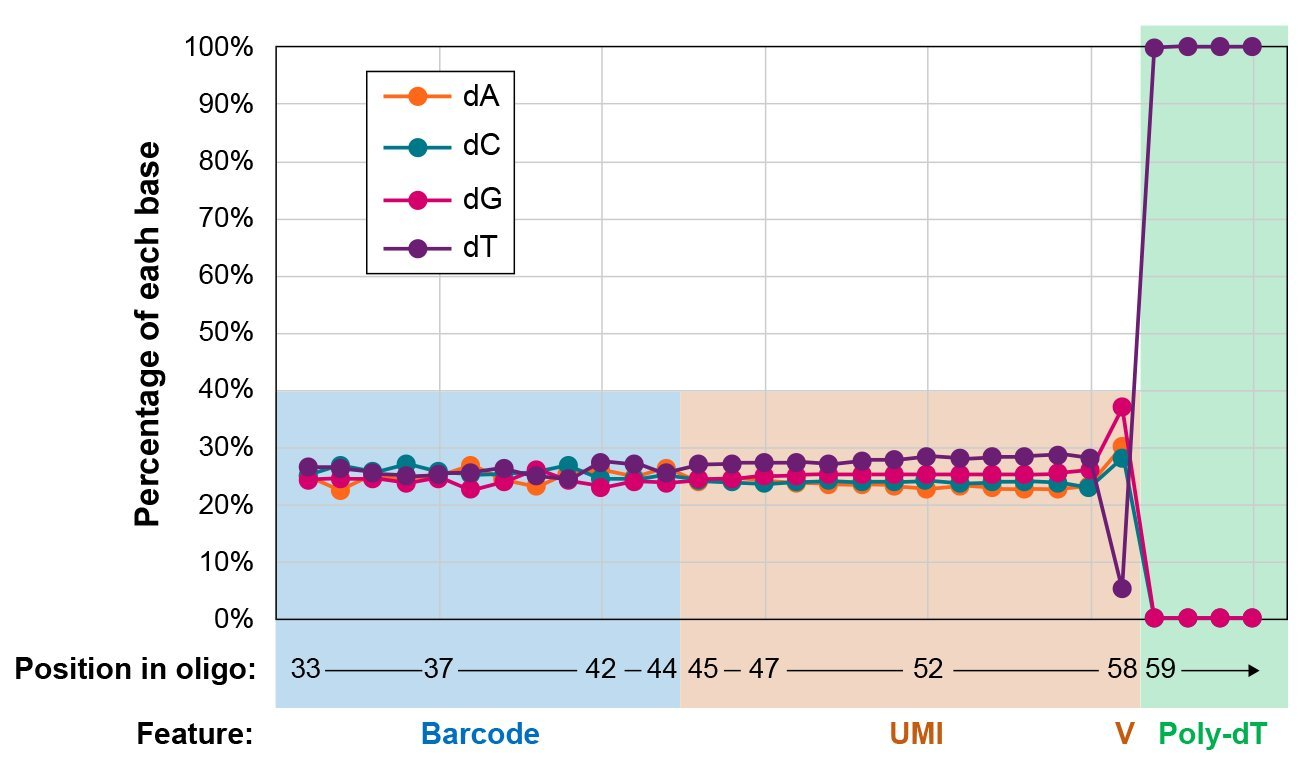
Figure 3. Typical sequencing results for the barcode, UMI and poly-dT portions of oligos within a pool of NxSeq Single-cell RNA-seq Beads. Note the nearly equal representation of each base within the bead barcode and UMI sequences, minimal dT present at the V position of the UMI demonstrating a lack of synthesis errors, and the complete absence of any non-dT bases within the poly-dT tail.
Functional testing by RNA capture identifies consistent, high performance lots
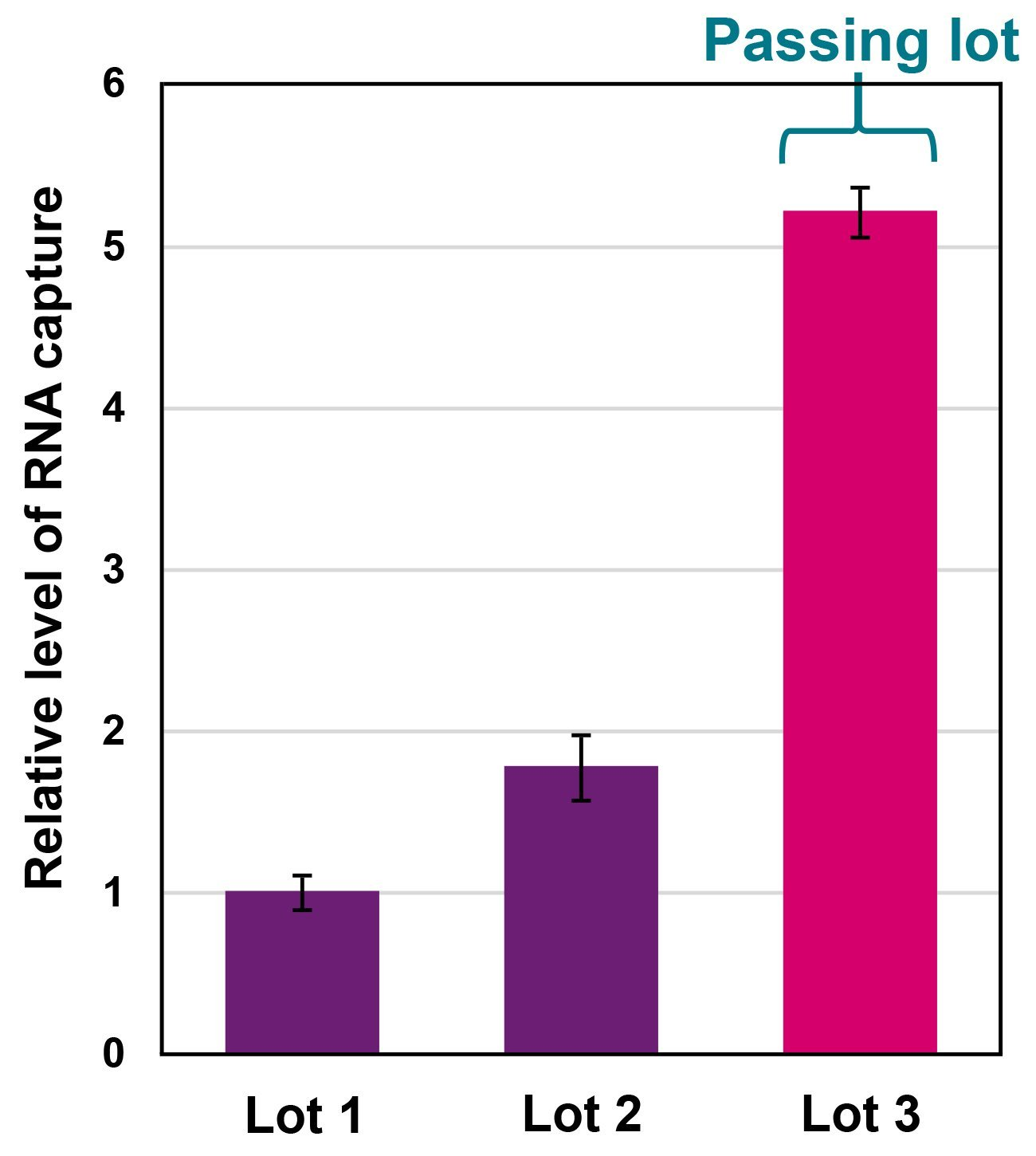
Figure 4. Example of RNA capture results done on different lots of NxSeq Single-cell RNA-seq Beads. In this assay, a poly-A RNA coupled to a DNA oligo (hybrid RNA) is spiked into total human RNA. The spiked RNA pool is hybridized to a pool of beads, washed, and the percent of captured hybrid RNA is measured by qPCR using a probe directed to the DNA sequence within the hybrid RNA oligo. Each of the three lots shown was tested in triplicate RNA captures and averaged. The average results per lot were normalised to Lot 1 and graphed.
References
1 E.Z. Macosko, A. Basu, R. Satija, J. Nemesh, K. Shekhar, M. Goldman, I. Tirosh, A. R. Bialas, N. Kamitaki, E. M. Martersteck, J. J. Trombetta, D. A. Weitz, J. R. Sanes, A. K. Shalek, A. Regev, S. A. McCarroll, Cell, 2015, 161, 1202-1214.
2 http://mccarrolllab.org/dropseq/




