Oligo Spacers: Your Flexible Friends
Please note: The description listed below includes a partial list of spacers currently available through LGC, Biosearch Technologies. The information is still accurate and may be used for general educational purposes. Prior to making a purchase please view the complete list of spacer products.
As with our amino and thiol linkers we are often asked why there is a need for so many spacer modifiers.
Spacers are, as their name suggests, designed to add space either within an oligo sequence or between the oligo (e.g. to add flexibility) and a label (e.g. FAM, biotin). This leads to another question we are often asked, how can the 3’-end of an oligo be modified with a spacer? Surely there must be something at either end of the ‘space’. In fact this is a very valid observation.
In this case, 3’-spacers such as LK2245 and LK2395 (See Figure 1) are often used as an alternative to 3’-phosphate as blockers since, when incorporated at the 3’-end, the resulting oligonucleotide shows nuclease and polymerase resistance. In fact spacer C3 is often incorporated at the 3’-end of an oligonucleotide for use with restriction enzymes rather than phosphate since this is thought to partially cleave during the assay.
Figure 1. 3’-Spacer Modifiers
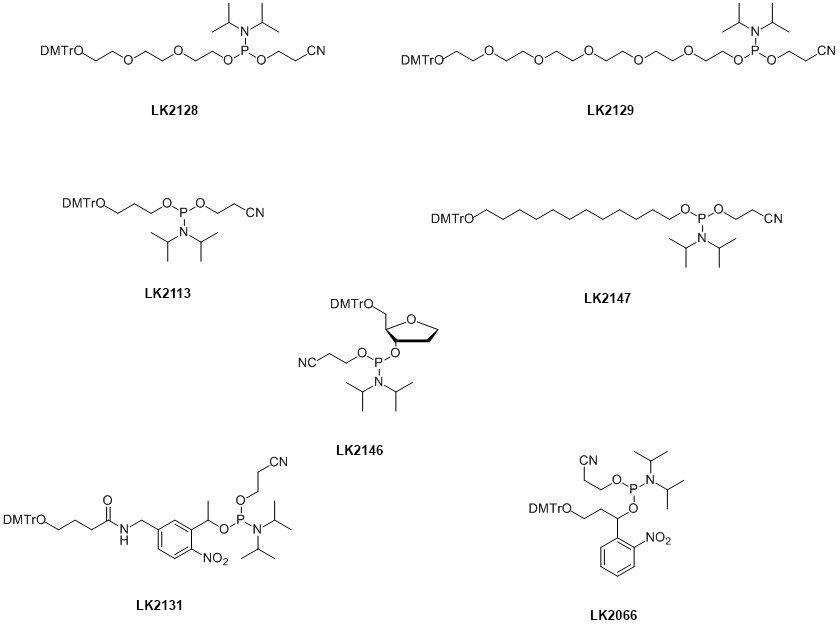
In addition to the 3’-‘spacers’ Link offers a variety of spacer phosphoramidites. These are shown in Figure 2 and are often used to distance a modifier from the oligonucleotide sequence.
This reduces the possibility of any adverse interaction between the modifier and the sequence. For instance, G-rich sequences are known to quench fluorescein therefore the use of a suitable spacer will remove the dye label from the proximity of the oligonucleotide minimising the quenching effect. In a similar fashion, spacers are often used to add distance between multiple additions of self-quenching dyes, e.g. fluorescein.1
The application of the modified oligonucleotide will dictate whether a hydrophilic (Spacer 18; LK2129, Spacer 9; LK2128) or hydrophobic spacer (Spacer C3; LK2113, Spacer C12; LK2147) is required.
Multiple incorporations of varying lengths of these spacers allow the precise length of the spacer arm to be controlled. This can be important in hairpin loop2 and duplex studies3 of DNA.
Several spacers have specific uses. Spacer C3 phosphoramidite (LK2113), when incorporated into an oligonucleotide, mimics the three carbon spacing between the 3’ and 5’ hydroxyls of sugar unit.4 Although useful where the base at a specific site is unknown, the flexibility of the alkyl chain distorts the sugar-phosphate backbone. This can be alleviated with the use of dSpacer (LK2146) since incorporation of this modifier sits directly into the natural sugar phosphate backbone with no adverse effect. This modifier mimics abasic sites5 and is useful in the study of mutations resulting from depurination.
Figure 2. Spacer Modifiers
The hydrophilic spacers; Spacer 9 (LK2128) and Spacer 18 (HEG) (LK2129) are also particularly useful to build in flexibility within an oligo sequence.6
As with 3’-‘spacers’ these are known to be nuclease and polymerase resistant which is why HEG is used in Scorpion primers® to not only add the flexibility required for the oligo to bend allowing hybridisation to the amplicon, it also acts as a PCR blocker preventing read through into the probe.7
Another example is Cytocell’s SMART detection assay.8 This uses spacer 18 or HEG (LK2129) in the template probe where one section acts as an anchor in binding to the target leaving the other section free for hybridisation to the extension probe to allow amplification during PCR. In this case the spacer gives flexibility to the template probe to enable hybridisation to both the target and the extension probe.
In some cases it is advantageous to remove the modifier from the oligonucleotide, For example, if using biotin as a means of capture, the target is bound to the biotin labelled probe. This is then captured using a streptavidin affinity column and the target-probe duplex can be eluted if a cleavable spacer is used; e.g. our photocleavable spacer LK2131.
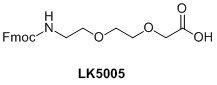
LGC, Biosearch Technologies also offers AEEA linker (LK5005); this can be used both as a spacer and a linker when used in PNA synthesis. Again this is used to distance a modifier from a PNA sequence or to add flexibility within a sequence, e.g. bisPNA.9
It is clear that there is a need for a variety of spacer modifiers to allow flexibility and reduce any adverse interactions between an oligo and a modifier. The application of the modified oligo will determine the type of spacer required. The amidites enable the possibility of incorporating a spacer or multiple additions of spacers anywhere within an oligo sequence. Similarly the AEEA linker (LK5005) plays the same role in PNA oligo synthesis. 3’-Spacers, despite their name, act as blockers rather than adding space or distancing a modifier from an oligo.
References
- See for example: Design of multidye systems for FRET-based applications, M.S. Shchepinov and V.A. Korshun, Nucleosides, Nucleotides & Nucleic Acids, 20, 369-374, 2001.
- Circular dichroism studies of an oligodeoxyribonucleotide containing a hairpin loop made of a hexaethylene glycol chain: conformation and stability, M. Durand, K. Chevrie, M. Chassignol, N.T. Thuong and J.C. Maurizot, Nucleic Acids Research, 18, 6353-6359, 1990. [Abstract]
- A nicked duplex decamer DNA with a PEG6 tether, L. Kozerski, A.P. Mazurek, R. Kawecki, W. Bocian, P. Krajewski, E. Bednarek, J. Sitkowski, M. P. Williamson, A.J.G. Moir and P.E. Hansen, Nucleic Acids Research, 29, 1132-1143, 2001. [Full Text PDF]
- Enhancing sequence-specific cleavage of RNA within a duplex region: Incorporation of 1,3-propanediol linkers into oligonucleotide conjugates of serinol-terpyridine, B.N. Trawick, T.A. Osiek and J.K. Bashkin, Bioconjugate Chem., 12, 900-905, 2001. [Abstract]
- (a) Oligodeoxynucleotides containing synthetic abasic sites model substrates for DNA-polymerases and apurinic apyrimidinic endonucleases, M. Takeshita, C.N. Chang, F. Johnson, S. Will and A.P. Grollman, J. Biol. Chem., 262, 10171-10179, 1987 [Abstract]; and (b) NMR-studies of abasic sites in DNA duplexes deoxyadenosine stacks into the helix opposite the cyclic analog of 2-deoxyribose, M.W. Kalnik, C.N. Chang, A.P. Grollman and D.J. Patel, Biochemistry, 27, 924-931, 1988. [Abstract]
- (a) Control of folding and binding of oligonucleotides by use of non-nucleotide linker, M. Salunkhe, T.F. Wu and R.L. Letsinger, J. Amer. Chem. Soc., 114, 8768-8772, 1992 [Abstract]; and (b) Oligonucleotide circularization by template-directed chemical ligation, N.G. Dolinnaya, M. Blumenfeld, I.N. Merenkova, T.S. Oretskaya, N.F. Krynetskaya, M.G. Ivanovskaya, M. Vasseur and Z.A. Shabarova, Nucleic Acids Res., 21, 5403-5407, 1993. [Abstract]
- (a) Mode of action and application of Scorpion primers to mutation detection, N. Thelwell, S. Millington, A. Solinas, J. Booth and T. Brown, Nucleic Acids Res., 28, 3752-3761, 2000 [Full Text PDF]; and (b) Duplex Scorpion primers in SNP analysis and FRET applications, A. Solinas, L.J. Brown, C. McKeen, J.M. Mellor, J.T.G. Nicol, N. Thelwell and T. Brown, Nucleic Acids Research, 29 (20), e96, 2001. [Full Text PDF]
- Detection of virus mRNA within infected host cells using an isothermal nucleic acid amplification assay: marine cyanophage gene expression within Synechococcus sp, S.D. Wharam, M.J. Hall and W.H. Wilson, Virology Journal, 4, 52-59, 2007. [Full Text PDF]
- An Experimental Study of Mechanism and Specificity of Peptide Nucleic Acid (PNA) Binding to Duplex DNA, H. Kuhn, V.V. Demidov, P.E. Nielsen and M.D. Frank-Kamenetskii, Mol. Biol., 286, 1337-1345, ,1999. [Full Text HTML]