Many next generation DNA sequencing applications or sample types require the construction of PCR-amplified DNA fragment libraries. (E.g. whole genome sequencing or resequencing from limiting genomic DNA amounts or FFPE and cell-free DNA samples, exome sequencing, ChIP-seq, etc.). To get the highest quality data for these applications and sample types, you need a DNA library preparation kit with the highest efficiency adaptor ligation followed by unbiased PCR amplification to produce the most complex, unbiased libraries possible. These high quality libraries produce the high quality sequencing data with uniform depth and minimal zero coverage needed to drive your NGS experiments forward.
The updated NxSeq™ UltraLow DNA Library Kits v2 and NxSeq Indexing Kits allow you to build high quality DNA fragment libraries from extremely low DNA input amounts – as low as 50 pg depending on sample type and Illumina sequencer used. If you have more DNA, no problem; you can use as much as 100 ng of input DNA with this system.
To generate these high quality libraries, we optimised each step of the protocol to ensure peak performance on Illumina sequencers. To help with the initial steps of DNA fragmentation, we have included guidance for mechanical shearing on an instrument like a Covaris LE220 and an optimised protocol for enzymatic fragmentation using dsDNA Shearase Plus from Zymo Research. In this updated kit, we replaced the PCR master mix with an improved master mix that performs better by decreasing bias across high AT-rich genomic regions. Not only will you produce high quality libraries, but also once you have your fragmented DNA, library prep is quick and easy - about three total hours.
Applications
These library and indexing kits are compatible with multiple applications and sample types including whole genome sequencing (WGS) and resequencing (WGR) applications for genomic assembly, SNV/mutation or copy number variation (CNV) identification. The system is also compatible with other NGS applications such as ChIP-seq and exome-seq. It also works well with both high quality input DNA samples as well as challenging sample types such as metagenomic, FFPE and cell-free DNA samples.
Formats
We offer 12 and 96 reaction kits with protocols to match both low throughput, tube-based library prep and high throughput, 96-well plate-based library prep. To complement these kits, we offer both single indexing and combinatorial dual indexing kits. If you are interested in using different indexing systems such as unique dual indexes or higher numbers of combinatorial dual indexes, we can help by providing you with information on the design of our indexing primers. With that information, you can order custom indexing primers that work well with our universal adaptor (Figure. 1 below). Please contact us for more information.
NxSeq UltraLow DNA Library Kit single index library construction
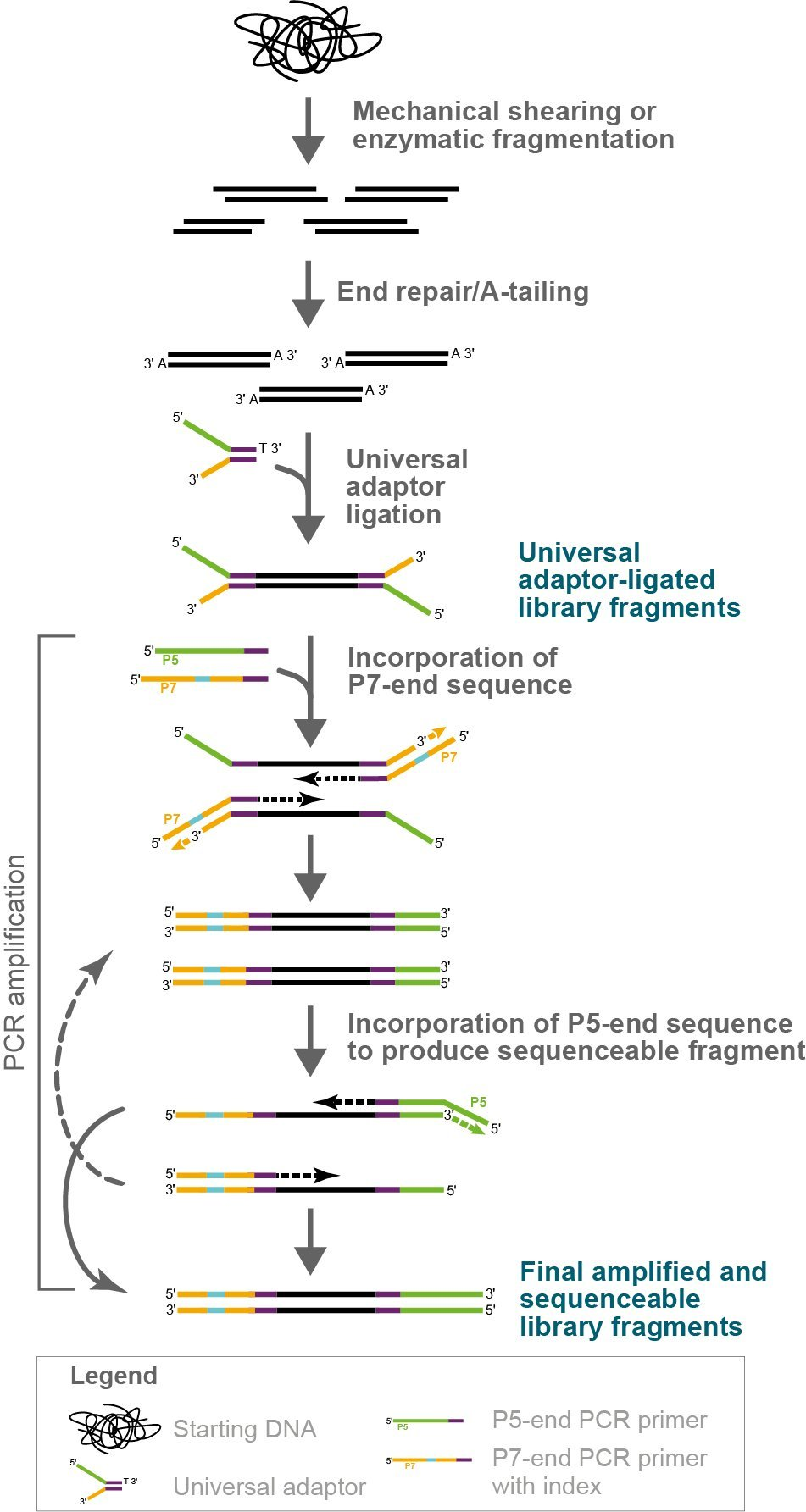
Figure 1. Mechanics of NxSeq UltraLow Kit library construction. This workflow figure illustrates how DNA fragment libraries are constructed using the NxSeq UltraLow DNA Library Kit v2 and NxSeq Single Indexing Kits. Dual indexed construction is the same, except both i5 and i7 indexes are incorporated during the PCR amplification step. Note that some PCR fragments and products are not shown to simplify the illustration.
Fast, streamlined library preparation with minimal components required
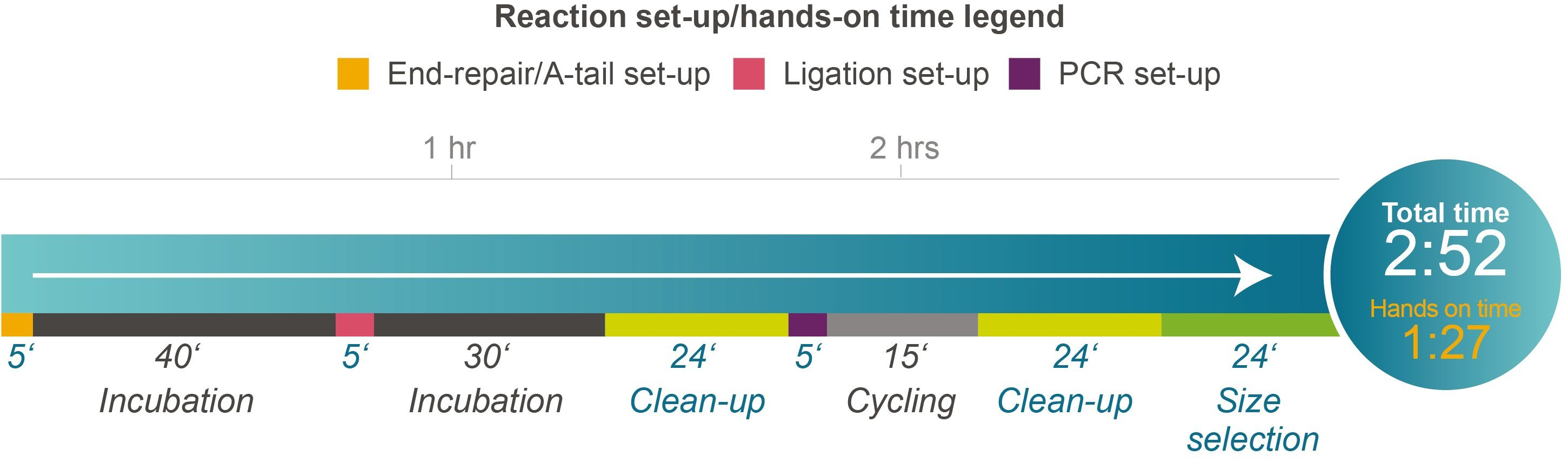
Figure 2. Time course for the NxSeq UltraLow DNA Library Kit v2 protocol. This figure illustrates the various steps and time required for each and total overall time. Please note that only three enzyme components are provided and required to complete library preparation, greatly simplifying library prep while reducing the risk of any errors.
Setting the stage for success with the high efficiency adaptor ligation
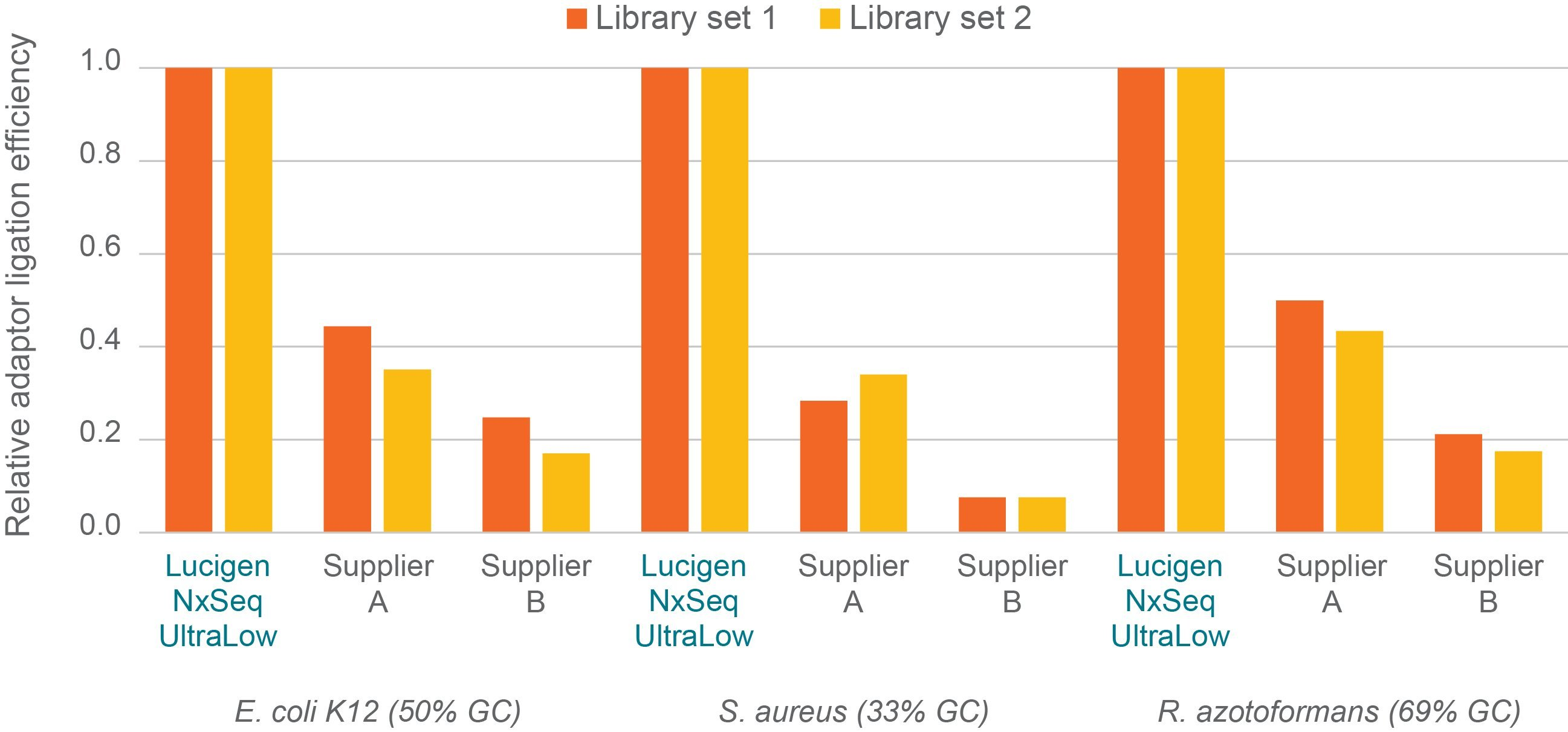
Figure 3. Efficiency of adaptor ligation to DNA library fragments. Two independent sets of libraries were prepped per kit/organism using manufacturer’s recommended protocols and 1 ng of the same sheared genomic DNA input per library. Briefly, DNA fragments with adaptors ligated to both ends were measured using triplicate qPCR assays and an universal qPCR primer set, designed by Biosearch Technologies, that binds to and amplifies all adaptor-ligated DNA fragments independent of the kit used. Efficiency was determined by comparing the qPCR quantitation to fluorescence DNA quantitation. Efficiency data was averaged and then normalised to the corresponding NxSeq UltraLow Library Kit data (1.0) and plotted.
Consistent, high quality sequencing data from 50 pg to 100 ng of genomic DNA
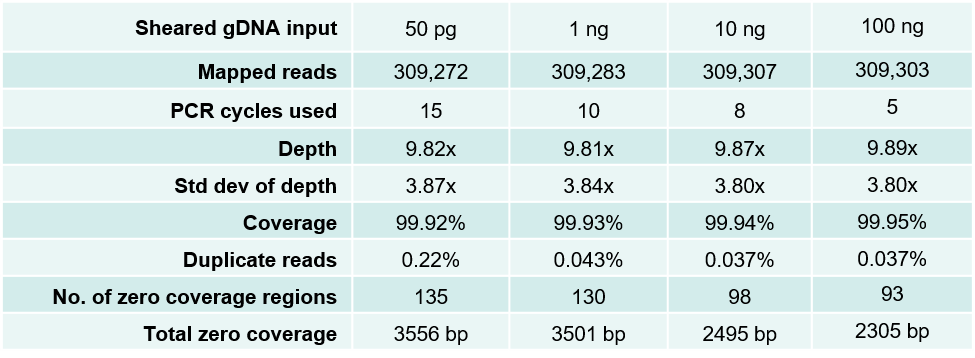
Table 1. Sequencing data from DNA fragment libraries generated from 50 pg to 100 ng of sheared E. coli gDNA. Mechanically sheared (Covaris LE220, peak size 300 bp) E. coli genomic DNA was serially diluted and used to make triplicate libraries starting with the indicated gDNA input amounts and number of PCR amplification cycles using the NxSeq UltraLow DNA Library Kit v2 and a NxSeq Single Indexing Kit. The final libraries were quantitated and diluted to 2 nM based on Bioanalyzer (size) and fluorescence-based DNA quantitation. The diluted libraries were then pooled and sequenced on a MiSeq using 2 x 150 bp chemistry, and the average data from the triplicate libraries are shown.
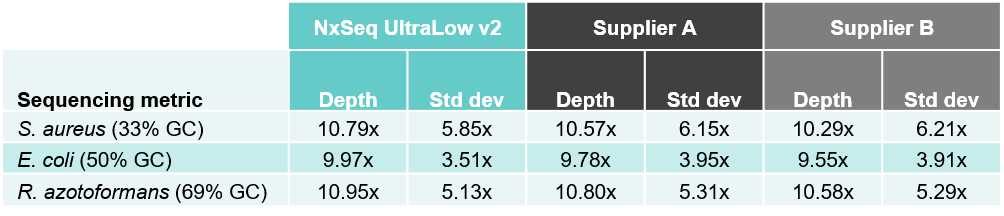
2A. Improved depth with less variation across three bacterial strains
2B. Better coverage – both fewer zero coverage regions and total zero coverage
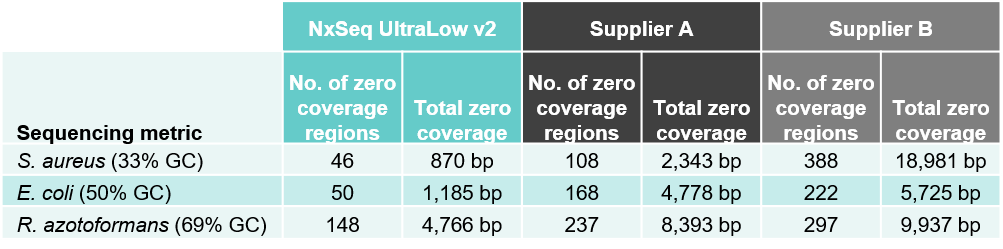
Table 2. Sequencing data from DNA fragment libraries generated from three bacterial species with varied genomic DNA GC content. Genomic DNA from S. aureus, E. coli, and R. azotoformans were mechanically sheared (Covaris LE220) to a peak size ~350 bp. Five ng of each sheared gDNA was used to create triplicate DNA fragment libraries using the NxSeq UltraLow Kit v2, and two other supplier kits according to manufacturer’s recommendations. Based on each user manual, the NxSeq libraries were amplified with 8 PCR cycles and Supplier A and B were amplified with 10 and 8 cycles, respectively. The final libraries were pooled and sequenced on a MiSeq using 2 x 150 bp chemistry. Equal numbers of mapped reads were analysed per library, and the average data from the triplicate libraries is shown. Table 2A compares the sequencing depth data from the three kits while Table 2B compares the zero coverage metrics.
Less GC- and AT-bias compared to other DNA fragment library kits
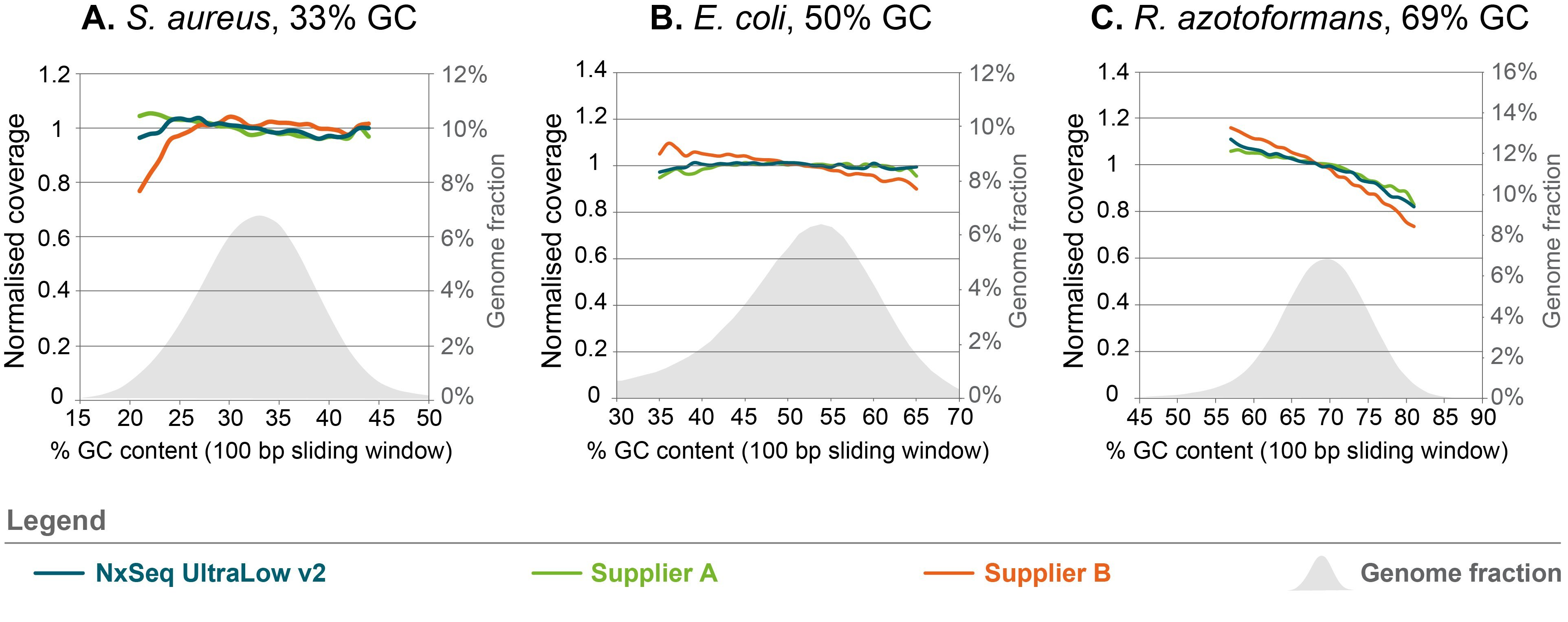
Figure 4. Sequence bias analysis for three different organisms with varying GC content. Triplicate genomic DNA fragment libraries were generated from gDNA of three organisms with varying GC content using the NxSeq UltraLow DNA Library Kit v2, Supplier A Kit or Supplier B Kit according to the manufacturer’s recommended protocols using 5 ng aliquots of the same mechanically sheared genomic DNA samples. The final libraries were pooled and sequenced on a MiSeq using 2 × 150 bp chemistry. Normalized coverage was calculated as the (average coverage of all windows with x% GC content) divided by the (overall average coverage) and the data from each set of replicates was averaged and presented. The grey shaded area represents the percent of each genome with the indicated GC content.
Highly consistent maize and human WGS results independent of gDNA input amount
A. Human results
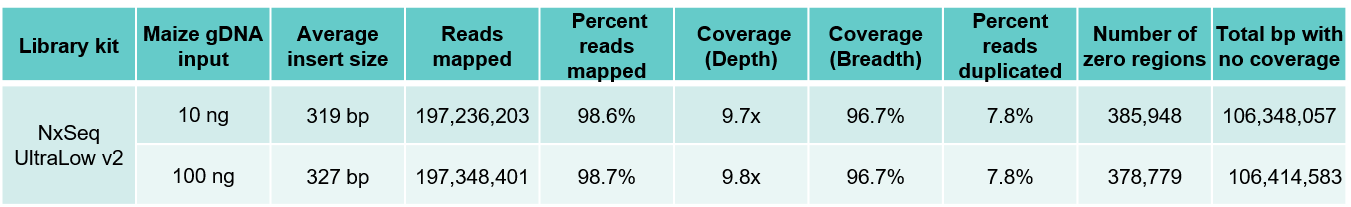
B. Maize results
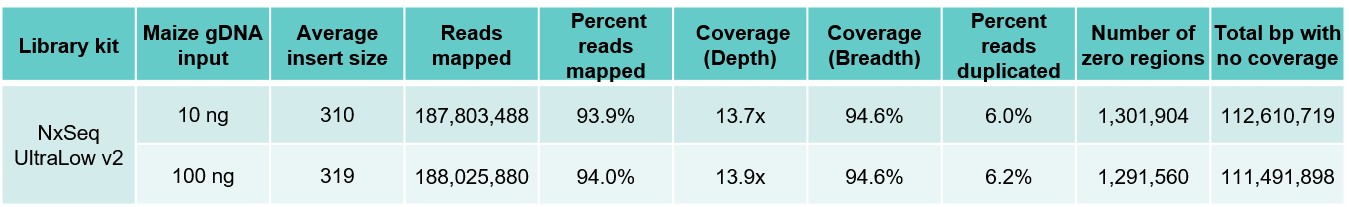
Table 3. Sequencing data from DNA fragment libraries generated from human (A) and maize (B) genomic DNA. Genomic DNA from human and maize B73 were mechanically sheared (Covaris LE220) to a peak size ~350 bp). Ten and 100 ng of each sheared gDNA was used to build triplicate DNA fragment libraries using the NxSeq UltraLow DNA Library Kit v2 as outlined in the user manual. The 10 and 100 ng libraries were amplified with 7 and 5 PCR cycles respectively. The final libraries were pooled and sequenced on a NovaSeq using 2 x 150 chemistry. Equal numbers of mapped reads (200M) were analysed per library, and the average data from the triplicate libraries shown.
Less GC- and AT-bias in maize libraries compared to other library kits
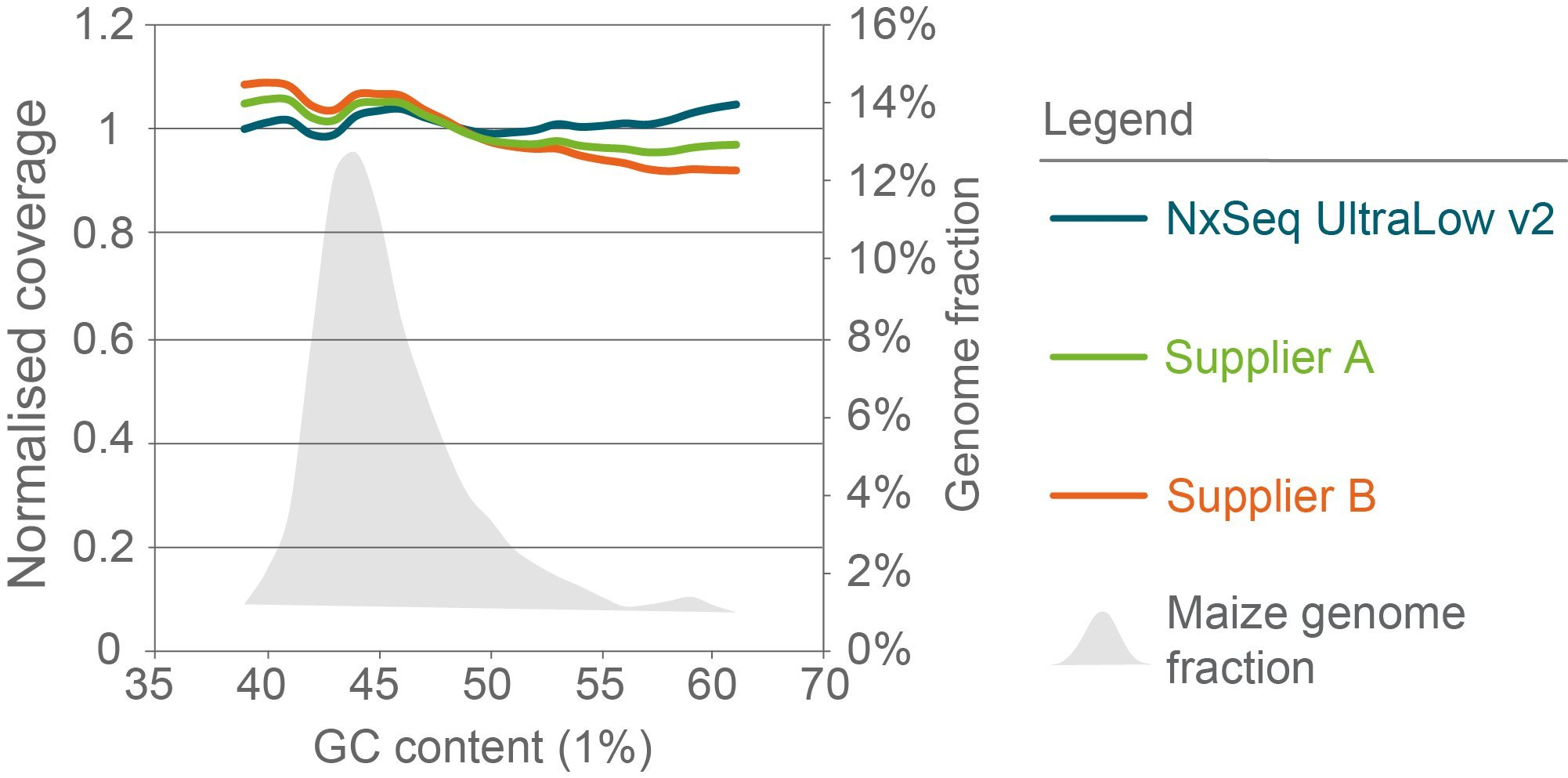
Figure 5
Sequence bias analysis
of maize libraries constructed with three library preparation kits. Triplicate genomic DNA fragment libraries were generated from maize B73 gDNA using the NxSeq UltraLow DNA Library Kit v2, Supplier A Kit or Supplier B Kit according to the manufacturer’s recommended protocols using 10 ng aliquots of the same mechanically sheared genomic DNA sample. The final libraries were pooled and sequenced on a NovaSeq using 2 × 150 chemistry. Normalised coverage was calculated as the (average coverage of all windows with x% GC content) divided by the (overall average coverage) and the data from each set of replicates was averaged and presented. The grey shaded area represents the percent of the maize genome with the indicated GC content.
Coloured dual indexing primer sets provide visual verification of correct primer dispensing
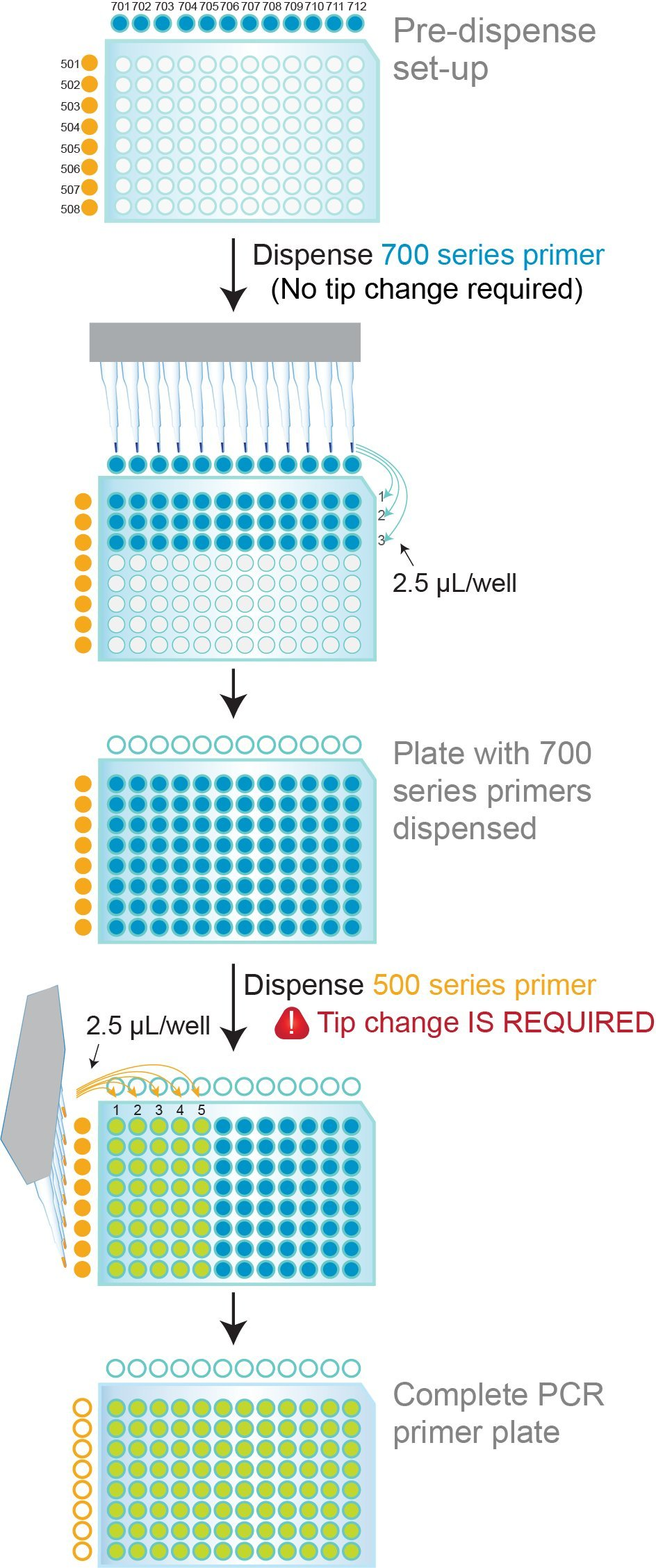
Figure 6. 96-well PCR plate NxSeq HT Dual Indexing Kit primer dispensing protocol. The 700 Series Indexing Primers in the NxSeq HT Dual Indexing Kit contain an inert blue dye and the 500 Series Indexing Primers contain an inert yellow/orange dye. The 700 Series Primers are dispensed first, row by row. As primers are dispensed, the rows become blue. The 500 Series Primers are then dispensed column by column, and as they are added, the well colour changes from blue to green. After dispensing is complete, all wells should exhibit a consistent green colour. The presence of clear, yellow/orange or blue wells indicates that those wells did not get the appropriate 700 and/or 500 Series Primers.










